Welcome! Use the links below to navigate:
Research Statement
I am always excited to work on open projects in algorithmic bioinformatics, evolutionary and population genetics. I develop scalable algorithms and pipelines for large-scale datasets, focusing on accelerating selection scans, k-mer–based compression, and analysis of sequence similarities. I also explore theoretical aspects of bioinformatics, including how tools interpret data and potential pitfalls in downstream analyses, to promote careful and accurate use of genetic and genomic data.
Bio
I am a postdoctoral scholar at Department of Biology, Pennsylvania State University, working with Dr. Zachary Szpiech on computational methods in population genetics. I obtained my PhD from Department of Computer Science and Engineering, Pennsylvania State University on August 2023. Before joining Penn State, I obtained my Bachelor’s degree in Computer Science and Engineering from Bangladesh University of Engineering and Technology (BUET). I also served as a lecturer in the CSE department at United International University, Bangladesh. During my PhD, I was a recipient of CBIOS Trainee Fellowship.
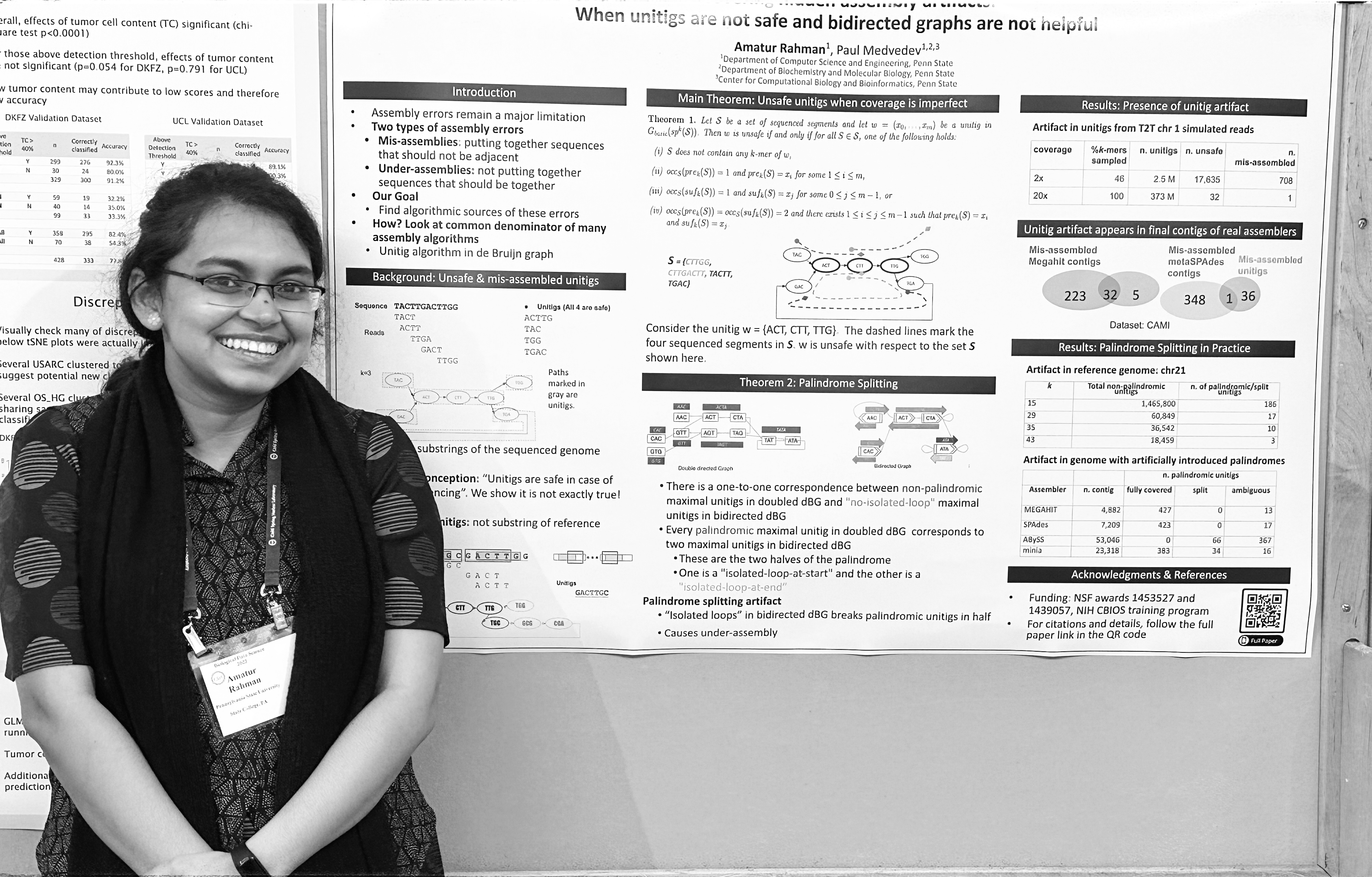
Biological Data Science Meeting, Cold Spring Harbor Laboratory, November 2022
CV
My most recent CV can be found here (last updated December 10, 2025).
Selected Publications
You can also check my publications through Google scholar.
-
Selection scans and downstream analysis with selscan.
Amatur Rahman*, T. Quinn Smith*, and Zachary A. Szpiech
bioRxiv, 2025-10. (Under revision at Human Population Genetics and Genomics)
(* joint first author)
[ paper ] -
Fast and memory-efficient dynamic programming approach for large-scale EHH-based selection scans.
Amatur Rahman, T. Quinn Smith, and Zachary A. Szpiech
Molecular Biology and Evolution, 42(11):msaf275, 2025.
[ paper ] -
Compression algorithm for colored de Bruijn graphs.
Amatur Rahman, Yoann Dufresne and Paul Medvedev
[ paper (preprint) ] -
Assembler artifacts include misassembly because of unsafe unitigs and under-assembly because of bidirected graphs.
Amatur Rahman and Paul Medvedev Genome Research, 32:1-8, 2022. An abstract appeared in RECOMB 2022, LNCS 13278:377–379 (Alternative title: Uncovering hidden assembly artifacts: when unitigs are not safe and bidirected graphs are not helpful.) [ paper (free version), paper (journal version) ] [talk] -
The K-mer File Format: a standardized and compact disk representation of sets of k-mers.
Y. Dufresne, T. Lemane, P. Marijon, P. Peterlongo, A. Rahman, M. Kokot, P. Medvedev, S. Deorowicz, and R. Chikhi Bioinformatics, 2022. [ paper ] -
GPU-accelerated and pipelined methylation calling.
Yilin Feng, Gulsum Gudukbay Akbulut, Xulong Tang, Jashwant Raj Gunasekaran, Amatur Rahman, Paul Medvedev, Mahmut Kandemir Bioinformatics Advances, Volume 2, Issue 1, 2022, [ paper ] -
Disk compression of k-mer sets.
Amatur Rahman, Rayan Chikhi and Paul Medvedev. WABI 2020 (accepted) Algorithms for Molecular Biology, 16(10), 2021. An extended abstract appeared in WABI 2020, LIPIcs 16:1–16:18. [ full version (journal), conference ] -
Representation of k-mer sets using spectrum-preserving string sets.
Amatur Rahman and Paul Medvedev.
Journal of Computational Biology, 28(4):381-394, 2021. An extended abstract appeared in RECOMB 2020, LNCS 12074:152-168. (Best Paper Award) [preprint] [talk] -
An Adaptive IoT Platform on Budgeted 3G Data Plans.
Mahmudur Rahman Hera, Amatur Rahman, Hua-Jun Hong, Li-Wen Pan, Md. Yusuf Sarwar Uddin, Nalini Venkatasubramanian, Cheng-Hsin Hsu. Elsevier Journal of Systems Architecture, 2018 -
Adaptive Sensing Using Internet-of-Things with Constrained Communications.
Mahmudur Rahman Hera, Hua-Jun Hong, Amatur Rahman, Pei-Hsuan Tsai, Afia Afrin, Md. Yusuf Sarwar Uddin, Nalini Venkatasubramanian, Cheng-Hsin Hsu. In Proceedings of Adaptive and Reflexive Middleware, ACM, Las Vegas, Nevada, USA, 2017 -
AQBox: An Air Quality Measuring Box from COTS Gas Sensors.
Mahmudur Rahman Hera, Amatur Rahman, Afia Afrin, Md. Yusuf Sarwar Uddin, and Nalini Venkatasubramanian. In Proceedings of 2017 International Conference on Networking, Systems and Security (NSysS), Dhaka, Bangladesh, 2017
Bioinformatics Software Development (Projects Led)
- selscan v3 – Fully redesigned and implemented EHH-based selection scans, achieving major improvements in speed, memory efficiency, and usability for downstream genomic analyses.
- ESS-Color – C++/Snakemake software for compressing colored de Bruijn graphs.
- ESS-Compress – C++ tool to compress a set of k-mers for disk storage.
- UST – C++ software to build a spectrum-preserving string set representation of k-mer sets.
Contact Info
Email: aur1111 “at” psu “dot” edu, amatur003 “at” gmail “dot” com